Publication
The paper for SparsePainter and PBWTpaint has now been published on Nature Communications: Yang, Y., Durbin, R., Iversen, A.K.N & Lawson, D.J. Sparse hap...
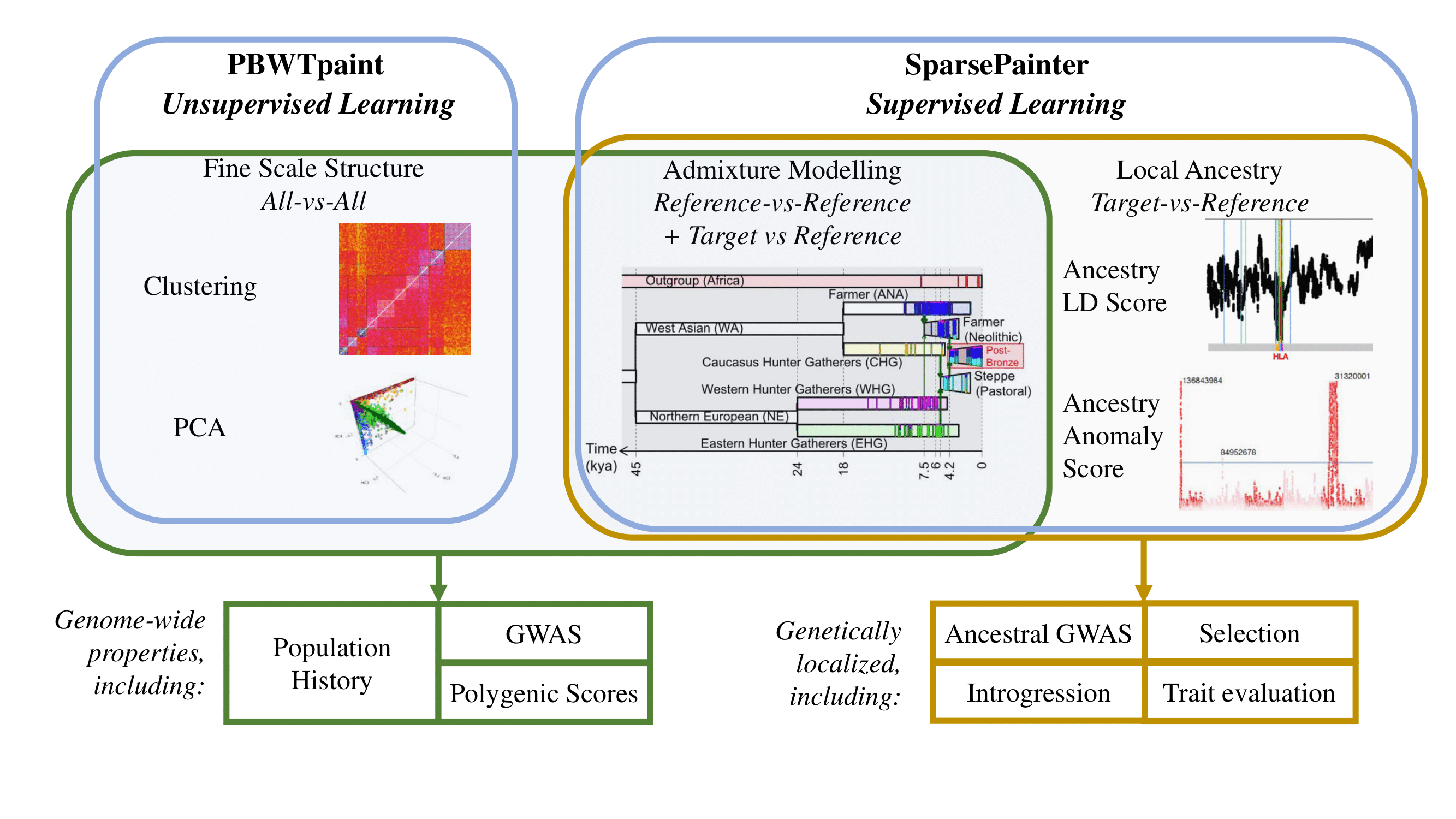
SparsePainter is an efficient tool for local ancestry inference (LAI) coded in C++. It extends PBWT algorithm to find K longest matches at each position, and uses the Hash Map structure to implement the forward and backward algorithm in the Hidden Markov Model (HMM) leveraging the sparsity of haplotype matches. SparsePainter can infer fine-scale local ancestry (per individual per SNP) and genome-wide total ancestry, it also enables efficiently calculating Linkage Disequilibrium of Ancestry (LDA), LDA score (LDAS) and Ancestry Anomaly Score (AAS) for understanding the population structure, evolution, selection, etc.
SparsePainter also produces output required to run GLOBETROTTER, fastGLOBETROTTER and SOURCEFIND (example codes to run SOURCEFIND based on SparsePainter output are available).
SparsePainter GitHub repository: https://github.com/YaolingYang/SparsePainter
PBWTpaint GitHub repository: https://github.com/richarddurbin/pbwt
SparsePainter and PBWTpaint Reference: Yang, Y., Durbin, R., Iversen, A.K.N & Lawson, D.J. Sparse haplotype-based fine-scale local ancestry inference at scale reveals recent selection on immune responses. Nature Communications 16, 2742 (2025).
LDA, LDA score and AAS reference and use cases: Barrie, W., Yang, Y., Irving-Pease, E.K. et al. Elevated genetic risk for multiple sclerosis emerged in steppe pastoralist populations. Nature 625, 321–328 (2024).
Pipeline for biobank-scale painting and computing haplotype components (HCs) are available.
PBWTpaint GitHub Repository: https://github.com/richarddurbin/pbwt
A use case of HCs: Yang, Y., Lawson, D.J. From individuals to ancestries: towards attributing trait variation to haplotypes. medRxiv (2025), doi: https://doi.org/10.1101/2025.03.13.25323895.
Authors:
Yaoling Yang (yaolingyang1998@gmail.com)
Daniel Lawson (dan.lawson@bristol.ac.uk)
Maintainer:
Yaoling Yang (yaolingyang1998@gmail.com)
Version: 1.3.2
SparsePainter is a direct improvement of ChromoPainter and is orders of magnitudes faster.
The detailed installation instructions, usages and commands, and examples are available.
The main usage of SparsePainter is to perform Target-vs-Reference painting for local ancestry estimates, and Reference-vs-Reference painting for admixture estimation. PBWTpaint has extraordinary performance in doing All-vs-All painting for clustering and learning haplotype principal components.
Here is an overview of the functionalities of SparsePainter and PBWTpaint.
The paper for SparsePainter and PBWTpaint has now been published on Nature Communications: Yang, Y., Durbin, R., Iversen, A.K.N & Lawson, D.J. Sparse hap...
Our preprint is now available online: Yang, Y., Durbin, R., Iversen, A.K.N & Lawson, D.J. Sparse haplotype-based fine-scale local ancestry inference at s...
Welcome to SparsePainter. You can find the introduction, installation instructions, Usages and commands and Examples.